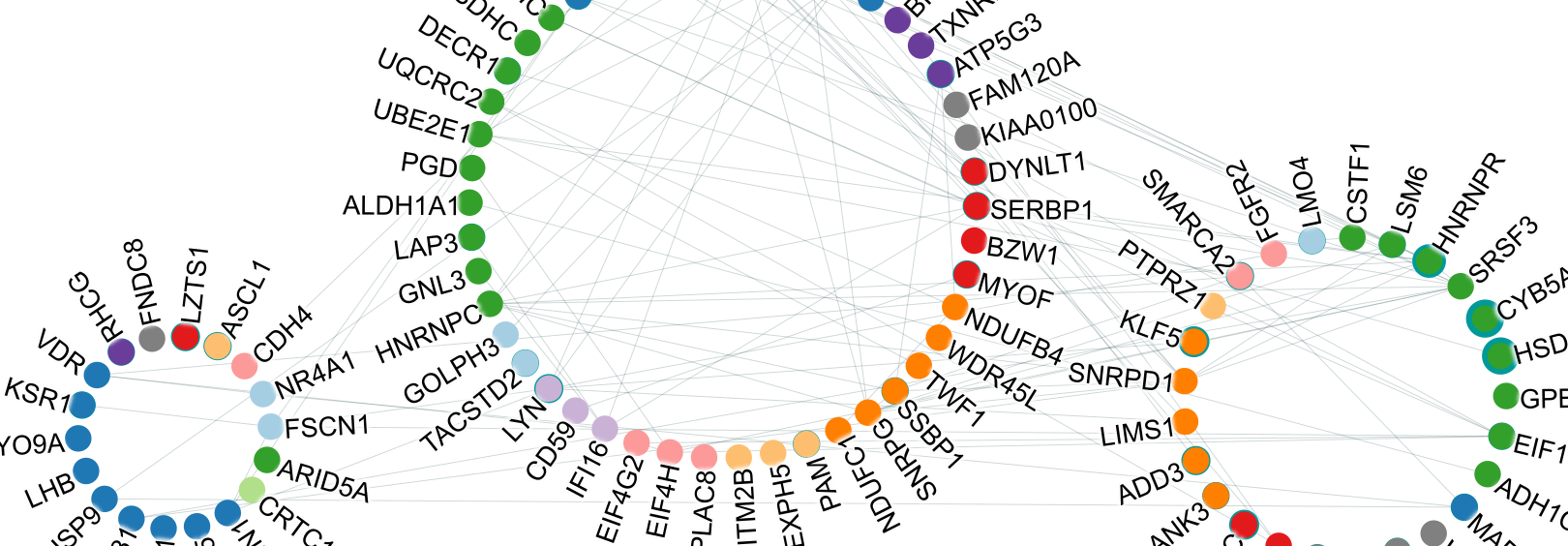
NAViGaTOR – Network Analysis, Visualization, & Graphing TORonto is an academic software package for visualizing and analyzing networks.
The current version, NAViGaTOR 3, implements a variety of functionality for network analysis, such as shortest path and clique search and static/dynamic layout algorithms such as circular or GRIP. Annotation of networks enable enrichment analysis of attributes discovering their distribution in the network. In particular the integrated protein protein network functionality enables the annotation of gene/protein with molecular function (e.g. Gene Ontology) or disease relevance (e.g. Human Disease Ontology).
Features
-
- NAViGaTOR combines a visualization system implemented in OpenGL with a graphical user interface and analytical algorithms implemented in Java.
-
- The NAViGaTOR workspace supports multiple network views (graph, spreadsheet, matrix, bar chart and serial data).
-
- Networks can be exported in tab-delimited, NAViGaTOR or XML formats, and multiple graphical formats (BMP, JPG, TIFF, SVG, PNG, GIF).
-
- Nodes can be positioned using a mixture of automatic force-directed layout and manual placement. One of the automatic layout algorithms used is a multi-level force-directed layout algorithm called GRIP (Graph dRawing with Intelligent Placement) [Gajer and Kobourov, 8th Symposium on Graph Drawing (GD), 2000]. Because of the nearly linear time complexity of GRIP, NAViGaTOR can visualize full interactomes from IID.
-
- Subnetworks that are highly interconnected, and nodes that are hubs (i.e. that interact with many other nodes), can be automatically identified to aid analysis.
-
- Subsets of a network can be bookmarked and manipulated using standard set operations, such as union, intersection and difference.
-
- The opacity of nodes and edges can be adjusted to change the contrast of elements in the network and emphasize selected elements.
Navigator is a research funded and devoted project under active development by members of Jurisica Lab of the Krembil Research Institute. The project was funded by the XandY research grand to further develop the functionality and enhance the interchangeability between Navigator and other tools (e.g. Cytoscape) for network analysis and visualization. Therefore, Navigator implements import and export functionality for the most common file formats, such as XGMML, PSI or text.
NAViGaTOR is freely-downloadable for academic and not-for-profit institutions, and can be installed and run on Microsoft Windows, Linux / UNIX, and Mac OS systems. NAViGaTOR is written in Java and uses JOGL (Java bindings for OpenGL) to support scalability, highlighting or suppressing of information, and other advanced graphic approaches.